Software
Phase-space-sampling 

Reduce a large dataset by strategically downselecting datapoints. The downselection preferentially keeps data points that have low probability and discards data points that are observed with high probability. The code can accommodate high-dimensions (we have tried up to 89 dimensions) and high number of data points (we have tried up to 100 millions). An MPI-parallel strategy is used to speed up the downselection. The probability estimation is done with a normalizing flow which can be trained with GPU to further accelerate training.
Using an iterative PDF estimation, the training cost does not increase with the number of datapoints, thereby allows reducing very large dataset at a reduced cost.
Starting from a large dataset non-uniformly sampled such as the left picture below. The probability density function of the dataset is estimated (picture below on the right).
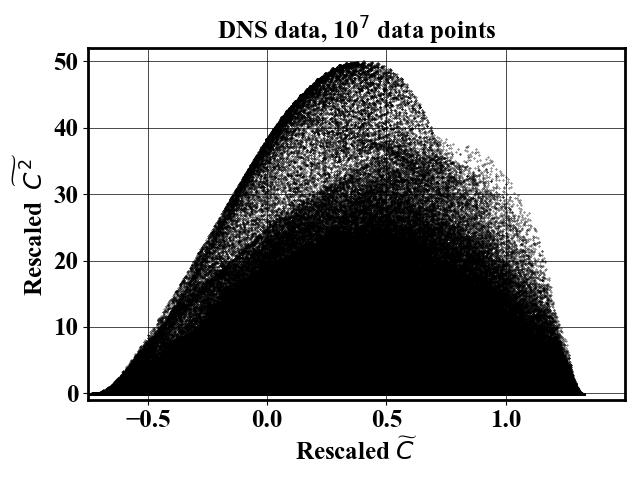
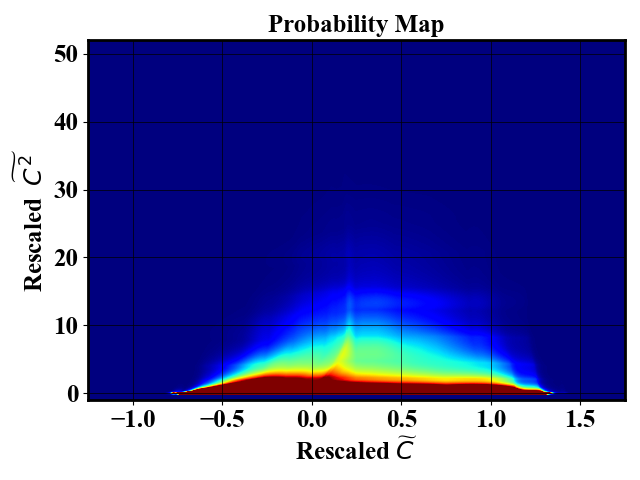
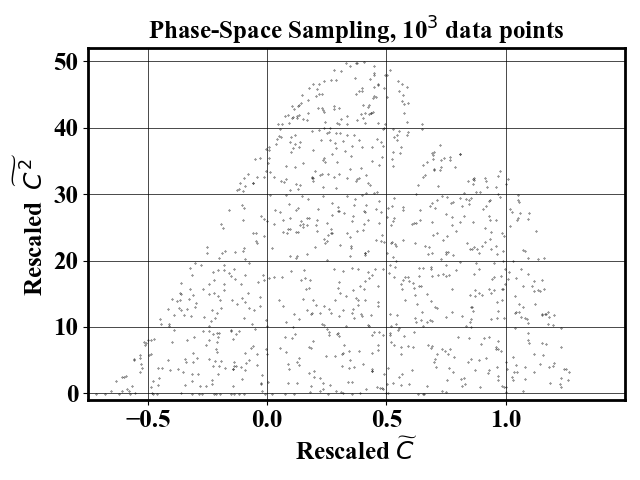
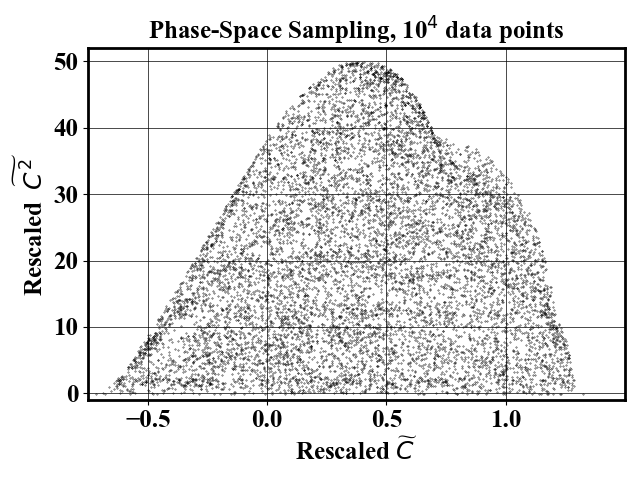
Uniform-in-phase-space datasets can be generated using the probability map using an arbitrary number of downselected points. The pictures below show a reduced dataset with 1,000 and 10,000 datapoints.
Bio Reactor Design (BiRD) 


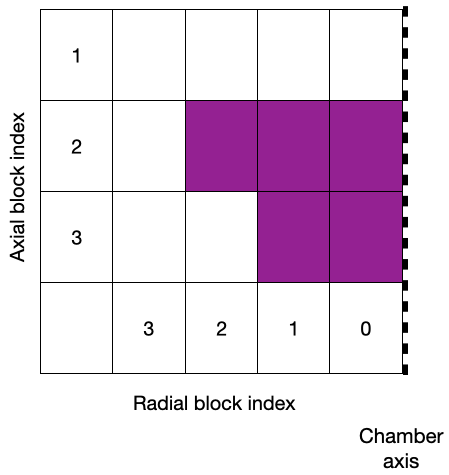
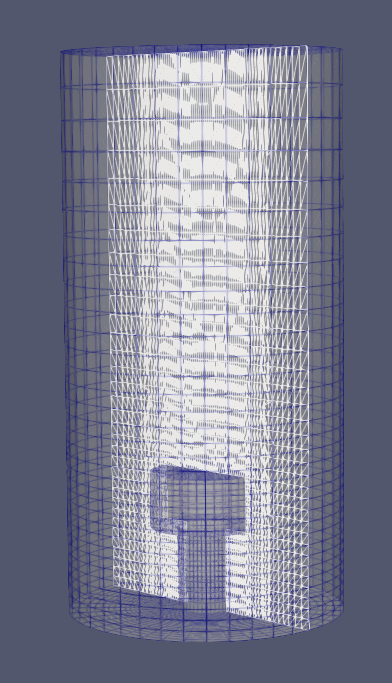
Generates clean meshes of block-cylindrical geometries. The code generates blockMeshDict which can be used to generate structured meshes with OpenFOAM. The interface to generate the blockMeshDict is designed to easily vary the geometry being meshed, thereby enabling geometry optimization tasks. A schematic of the block cylindrical architecture described by .json files in the repo is show below (left). The corresponding mesh generated is shown on the right.
Another interface is provides to generate STL files which may be used to generate boundary conditions in OpenFOAM. An example of such STL is shown below.
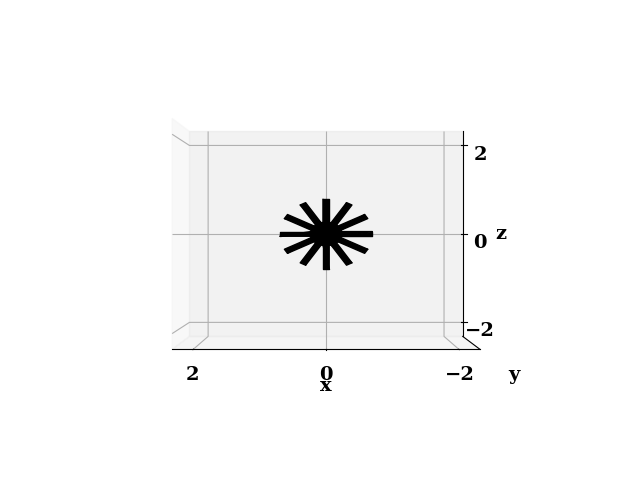
This code was primarily designed to optimize the design of spargers in bubble column reactors. However we can use it for any block cylindrical geometries which come up a lot more often than I initially thought!
Sup3r 


Super-resolution of global atmospheric models with Generative Adversarial Networks (GAN). The super-resolution may be spatial, temporal or both. The code is designed to handle very large datasets and large models. An interface to estimate the uncertainty during the super-resolution process is also provided.
Diversity Super Res
Diverse super-resolution of atmospheric data using a GAN (Generative Adversarial Neural Net). The diversity of the data generated is evaluated by computing a priori the conditional mean \(\mathbb{E}(HR \| LR)\) and the diagonal of the conditional variance \(\mathbb{E}(HR^2 \| LR)\). Examples of the a priori estimated moments are shown below.
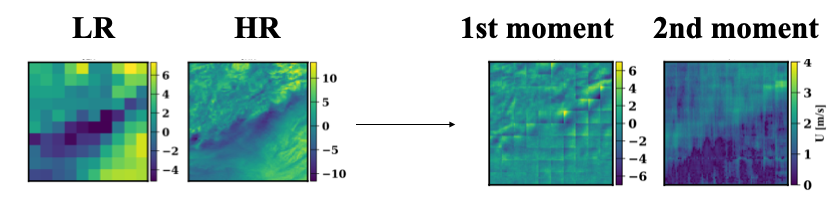
During training, minibatches of atmospheric data are generated to evaluate empirically the moments aforementioned for the generated samples. The a priori and empirical moments are compared through a Frechet Distance loss which penalizes the generator. The advantage of this method is that the distribution of the generated samples can be quantitatively compared to the ideal distribution.
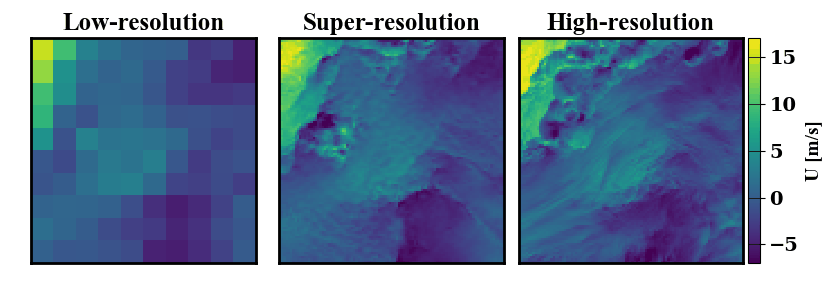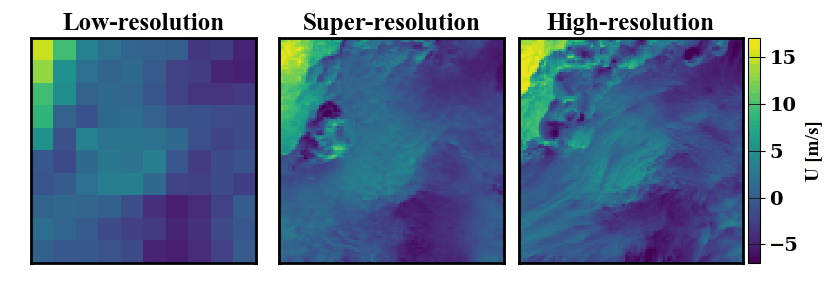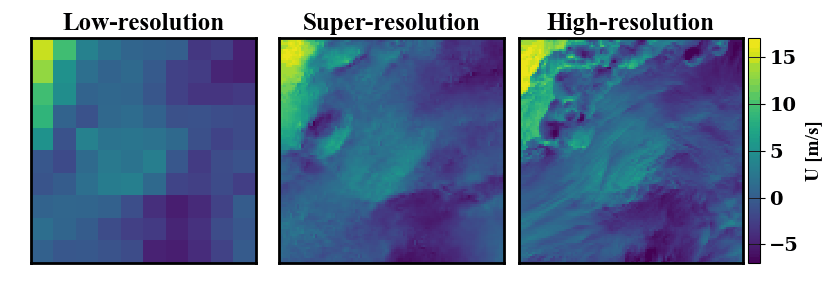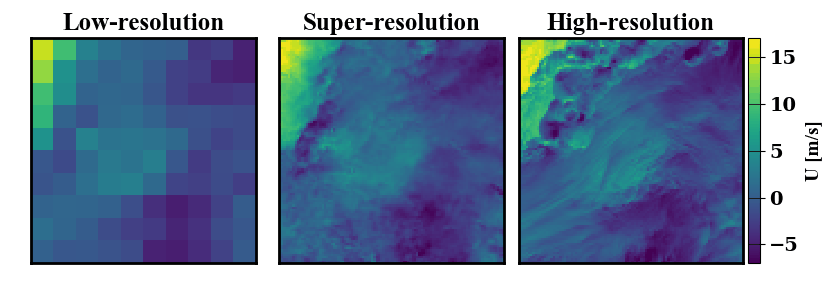
Below is a demonstration of the data that can be generated. On the left a low-resolution wind data input is shown, on the right the true high-resolution data is show. In the middle, many possible super-resolved wind contour that map back to the same low-resolution input are shown.
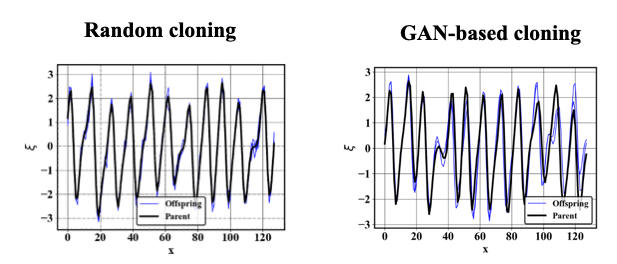
GANISP (GAN-assited Importance SPlitting)
Regularize genealogical adaptive multilevel splitting methods by using a Generative adversarial network (GAN) for the realization cloning. The GAN is regularized to encourage diverse generation of samples. Example of generated clones using the GAN vs a random cloning method are shown below in the case of Kuramoto-Sivashinsky equation.
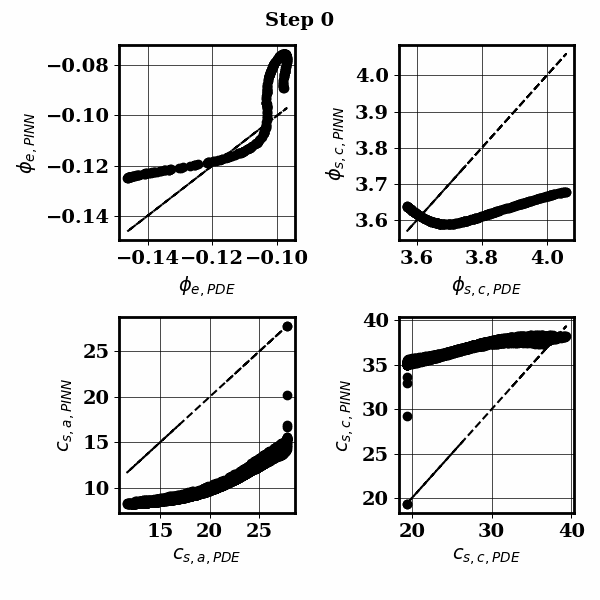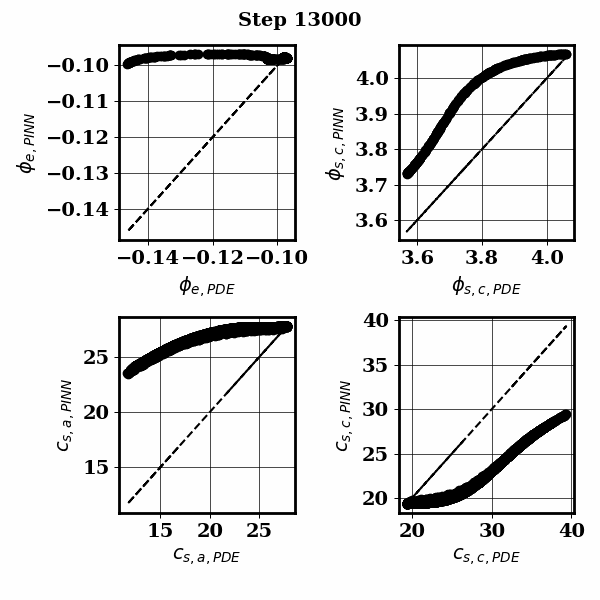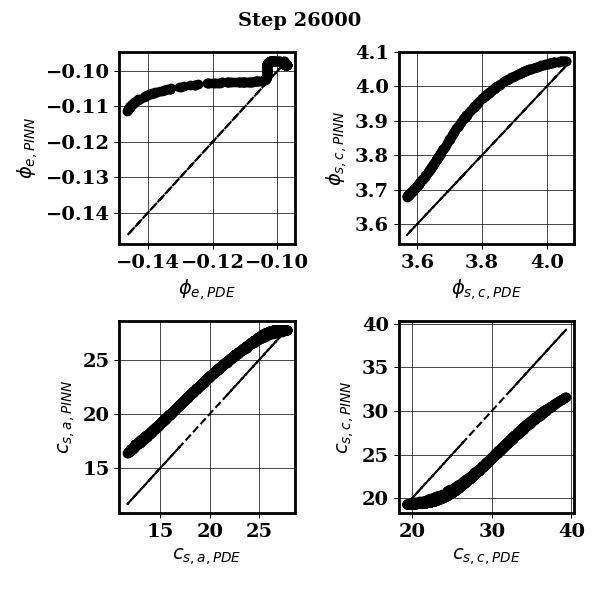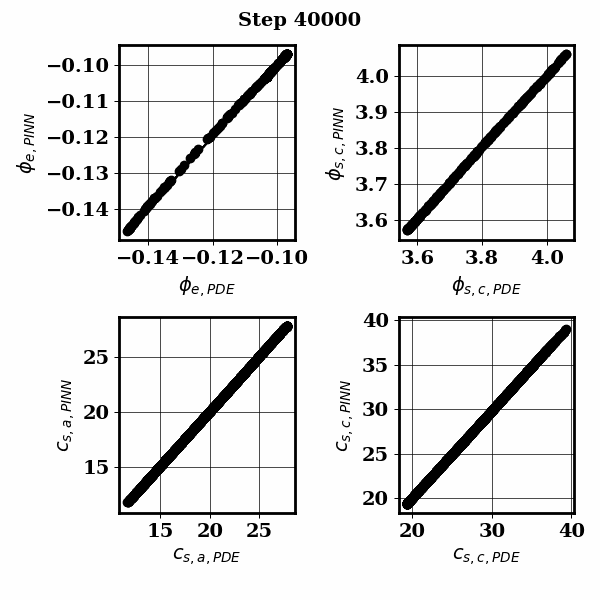
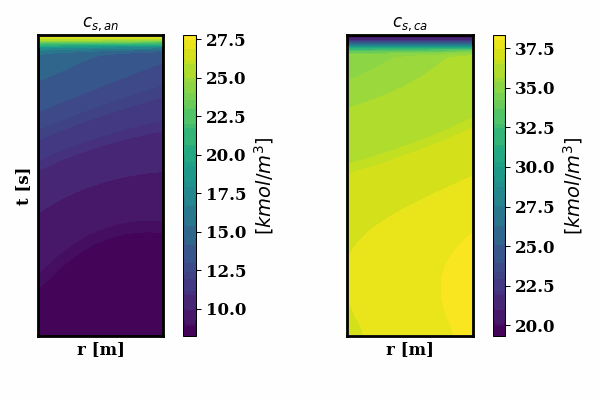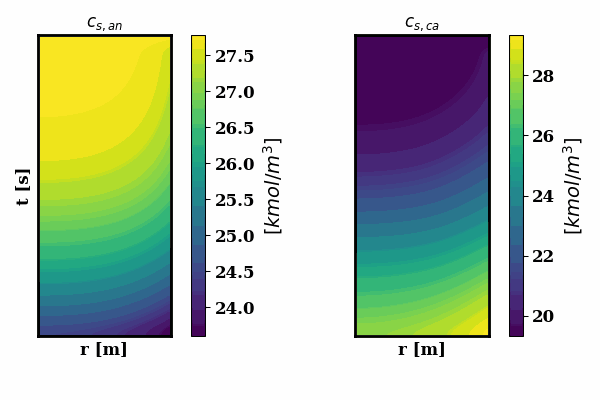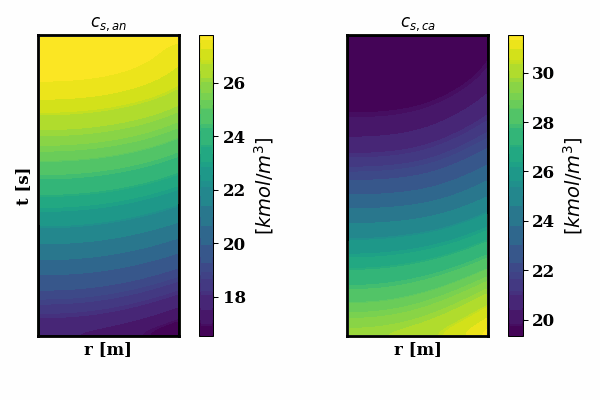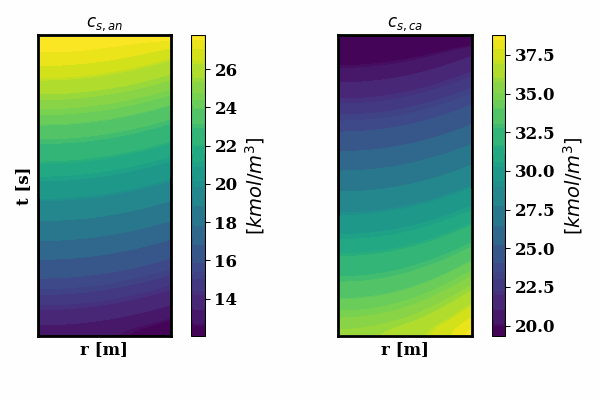
PINNSTRIPES (Physics-Informed Neural Network SurrogaTe for Rapidly Identifying Parameters in Energy Systems) 
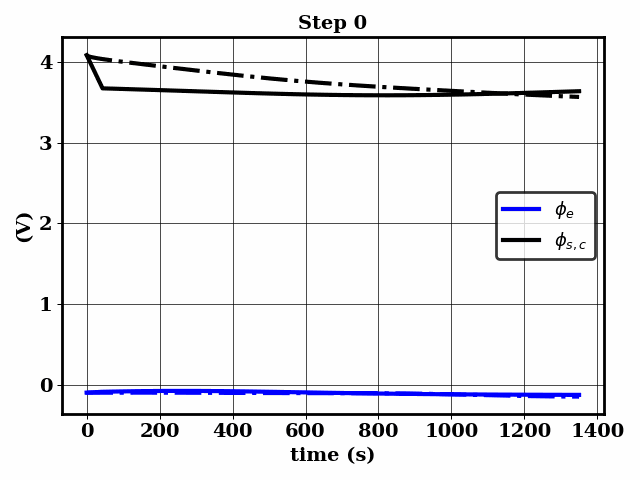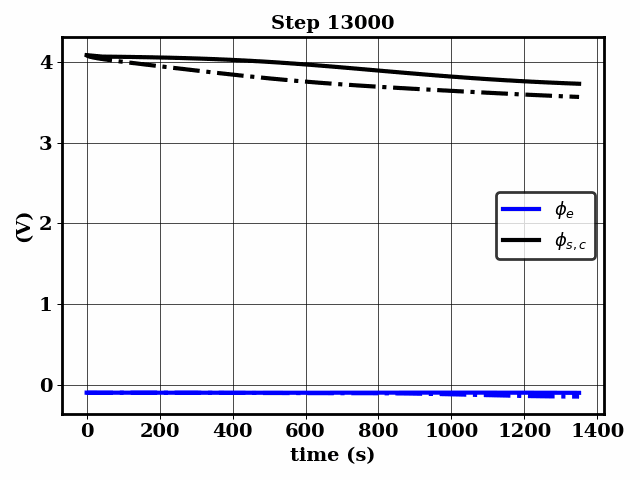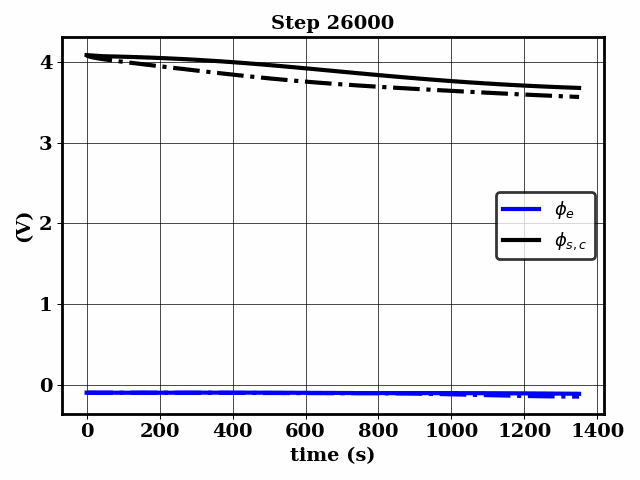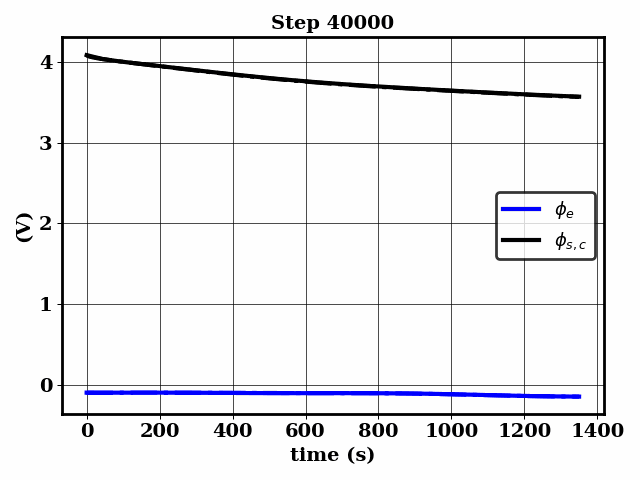
Bayesian inference of Li-ion battery models parameters made data-efficient thanks to Physics-informed neural networks. The code contains a demonstration of the implementation for single particle model, and provides an interface for using the surrogate within a Bayesian inference worflow.
As example the training results using no data is shown for the single particle model (SPM) of an NMC Li-ion battery.
prettyPlot 

A suite of tools to
- Standardize research plots in python
- Parse input files
- Display a progress bar for iterative processes
